Our work
PRODRUGS OF ANTIVIRAL NUCLEOTIDE ANALOGUES
Prophylactically administered USC-087 is more effective in counteracting the effects of disseminated Ad6 infection in female Syrian hamsters than equimolar doses of CDV.
USC-087 was administered p.o. starting at one day prior to infection and continued daily for the course of the experiment. CDV was administered i.p. starting with an induction dose one day before virus challenge and continued with 3 weekly injections for the duration of the experiment. A. Diagram of the experimental design. B. Survival. Ad6-Vehicle v. Ad6-USC-087 1 mg/kg p=0.0047; Ad6-Vehicle v. Ad6-USC-087 3 mg/kg or Ad6-USC-087 10 mg/kg p<0.0001; Ad6-Vehicle v. Ad6-CDV 0.8 mg/kg p=0.4505; Ad6-Vehicle v. Ad6-CDV 2.4 mg/kg p=0.0178; Ad6-Vehicle v. Ad6-CDV 8 mg/kg p=0.0001; Ad6-Vehicle v. Ad6-CDV 8 mg/kg p<0.0001 (Log rank); C. Serum alanine transaminase levels. Empty symbols in this and subsequent graphs indicate that the sample was collected from a moribund animal sacrificed ahead of schedule. D. Virus burden in the liver. *: p<0.05, **: p<0.01; for the purposes of statistical calculations, a value of 5x104 TCID50/g was assumed for all not detectable and not quantifiable samples; NQ: not quantifiable; ND: not detectable.
USC-087 reduces mortality and morbidity even when administered 3 or 4 days after virus challenge.
CP-treated hamsters were infected i.v. with Ad6 and left untreated or treated with USC-087 at 10 mg/kg p.o. q.d. Treatment with the drug started at 1 day before or 1, 2, 3, or 4 days after virus challenge. A. Diagram of the experimental design. B. Survival. n=15. Ad6-Vehicle v. all Ad6-USC-087 p<0.05 (Log rank) C. Serum ALT levels at 7 days post challenge n=5 to 9. *: p<0.05 D. Virus burden in the liver at 7 days post challenge. Empty symbols represent data collected from moribund animals. **: p<0.01, ***: p<0.001; for the purposes of statistical calculations, a value of 5x104 TCID50/g was assumed for all not detectable and not quantifiable samples), NQ: not quantifiable, ND: not detectable. E. Mean body weight changes for the Vehicle-Vehicle and the Vehicle-USC-087 groups. p<0.0001 (Two-Way ANOVA).
NUCLEOTIDE ANALOGUES TO STUDY DNA & RNA POLYMERASE
Synthesis of Individual α,β-CHX (X = F, Cl) ATP Diastereomers
Enzymatic approach to resolve a mixture of α,β-CHX-ATP diastereomers.
Utilization of γ-S-ATP, (R)-α,β-CHF-γ-S-ATP and (S)-α,β- CHF-γ-S-ATP by various kinases to phosphorylate substrate proteins in cell lysates. All γ-S nucleotides were used at 0.25 mM, and thiolphosphorylation was detected using a monoclonal antibody.
IMAGING PROBES FOR DRUG DEVELOPMENT
TREATMENT OF HEARING LOSS
Synthesis of compound RIS-DHF.
Spiral ganglion neurite outgrowth in vitro.
Immunohistochemical analysis of cochlear SGNs after 48 h in culture, treated with 400 nM DHF, 400 nM Ris, 400 nM Ris-DHF, or DMSO as a control. Neurons were stained with neural marker TuJ1 (red); nuclei are labeled with DAPI (blue). Scale bar represents 100 μm. Neurite outgrowth length was measured with 3D Neurite Tracer, and total traces were processed and rendered into a tagged 3D skeleton.
Prebinding to hydroxyapatite bone matrix does not inhibit 400 nM Ris-DHF-promoted neurite outgrowth in vitro.
∼500 μg HA nanoparticles prebound to 400 nM of Ris, DHF, Ris-DHF, or DMSO control were plated with SGNs for 48 h. Total traces were processed and rendered into a tagged 3D skeleton (white).
SELECTIVE INHIBITION OF FUNGAL BROMODOMAINS
Synthesis of compound 3.
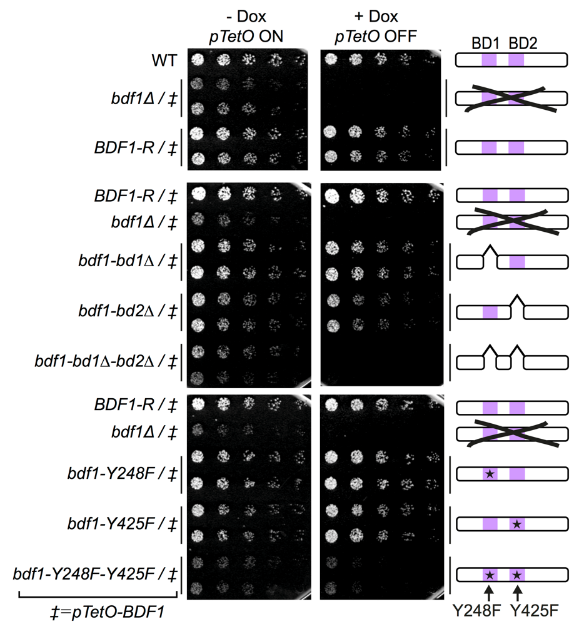
Bdf1 BDs are essential for C. albicans viability and virulence.
Colony formation assays showing the effect of Bdf1 repression and Bdf1 BD inactivation on growth. This experiment was repeated three times with similar results.
A CaBdf1 BD1 inhibitor phenocopies the effect of BD1 inactivation on C. albicans growth.
A. Chemical structure of compound 3. B. HTRF assay showing that 3 inhibits CaBdf1 BD1 preferentially over CaBdf1 BD2 and with modest selectivity over Brd4 BD1. C. Representative ITC experiments showing the binding of 3 towards the indicated BD. The values indicated for Kd and N represent the mean and s.d. from three independent experiments. See also Supplementary Table 1. D. Crystal structure of CaBdf1 BD1 (violet) bound to 3 (S enantiomer) and alignment with CaBdf1 BD1 bound to 1 and with human Brd4 BD1 (green). Side chains are shown for CaBdf1 residues in contact with 3 and for the corresponding Brd4 residues if divergent from CaBdf1. Inset, simulated-annealing omit Fo-Fc density for compound 3 contoured at 3σ.